…clip data to the extent of a vector file?#
The utility function mask_from_vec can be used to create a boolean mask from a vector
file, which can then be applied to an xarray.DataArray or xarray.Dataset object.
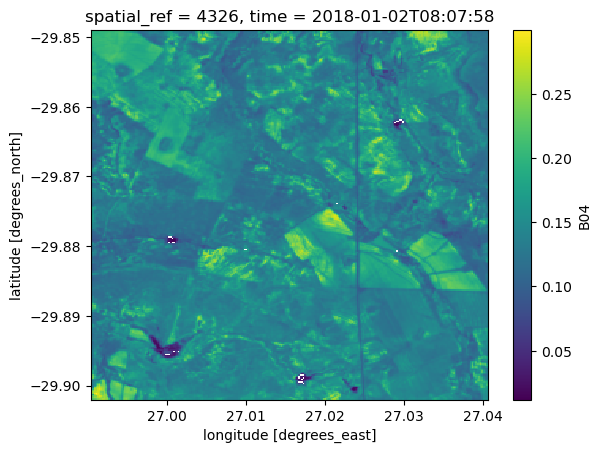
In the following example, we load Sentinel-2 L2A data using a vector file that contains a multipolygon. When we plot a single time step, we notice that the data is by default loaded for the bounding box of the vector file and not the multipolygon itself:
from sdc.load import load_product
ds = load_product(product='s2_l2a', vec=vec,
time_range=("2018-01-01", "2022-12-01"))
ds.B04.isel(time=0).plot()
<matplotlib.collections.QuadMesh at 0x7fe69ccb6840>
Before using the mask_from_vec-function, let’s first have a look at its docstring to
understand what it does and what it needs as input. We can do this by typing a question
mark followed by the function name:
import sdc.utils as utils
utils.mask_from_vec?
Signature:
utils.mask_from_vec(
vec: str,
da: Optional[xarray.core.dataarray.DataArray] = None,
) -> numpy.ndarray
Docstring:
Create a boolean mask from a vector file. The mask will have the same shape and
transform as the provided DataArray. If no DataArray is given, the `sanlc` product
will be loaded with the bounding box of the vector file and used as the template.
Parameters
----------
vec : str
Path to a vector file readable by geopandas (e.g. shapefile, GeoJSON, etc.).
da : DataArray, optional
DataArray to use as a template for the mask, which will be created with the same
shape and transform as the DataArray. If None (default), the `sanlc` product
will be loaded with the bounding box of the vector file and used as the
template.
Returns
-------
mask : ndarray
The output mask as a boolean NumPy array.
Examples
--------
>>> import sdc.utils as utils
>>> from sdc.load import load_product
>>> vec = 'path/to/vector/file.geojson'
>>> ds = load_product(product='s2_l2a', vec=vec)
>>> mask = utils.mask_from_vec(vec=vec, da=ds.B04)
>>> ds_masked = ds.where(mask)
File: ~/0000_sdc-tools_development/sdc-tools/sdc/utils.py
Type: function
It only requires a vector file (the one we have already used for loading) and optionally
a reference DataArray to determine the shape and transform of the output mask. We can
just use any band of the dataset we want to apply the boolean mask to.
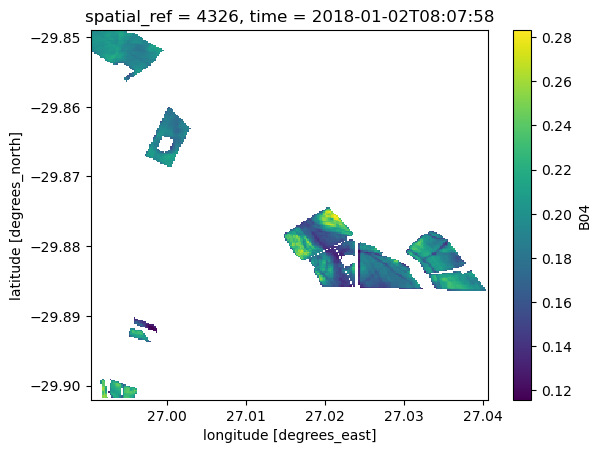
mask = utils.mask_from_vec(vec=vec, da=ds.B04)
ds_masked = ds.where(mask)
ds_masked.B04.isel(time=0).plot()
<matplotlib.collections.QuadMesh at 0x7fe69cca5b80>